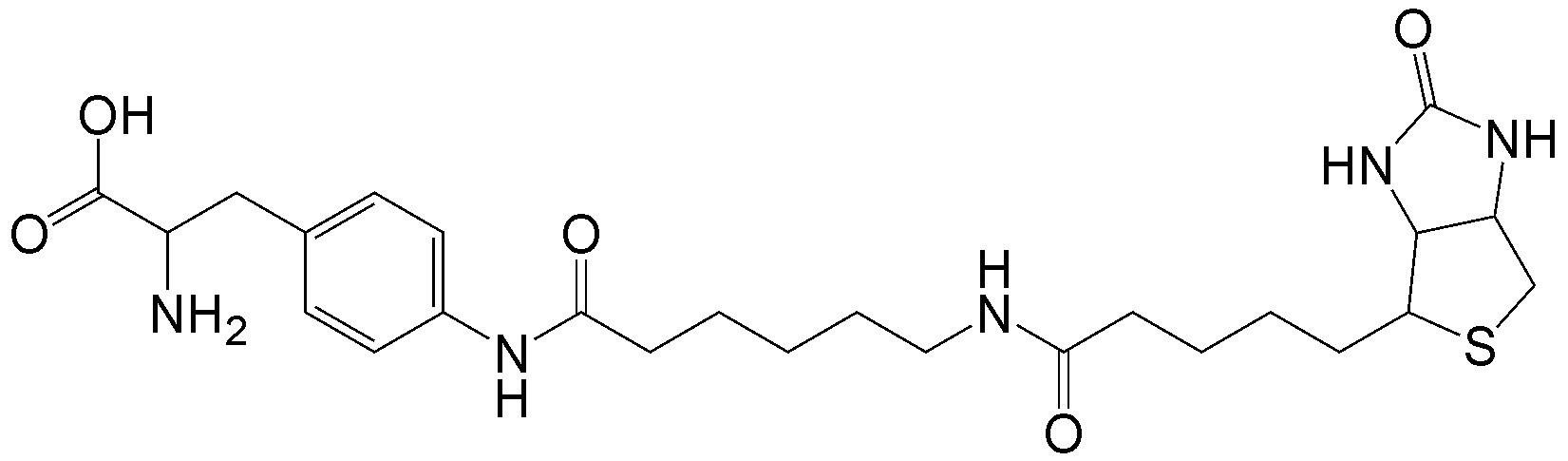
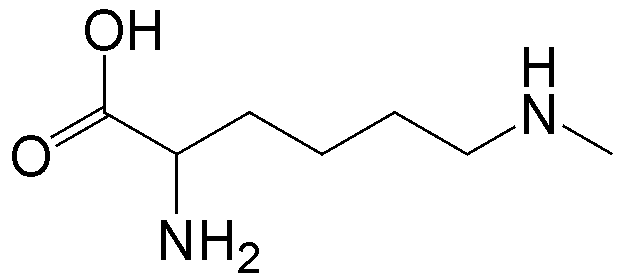
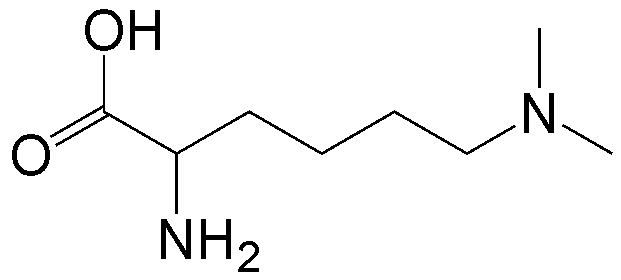
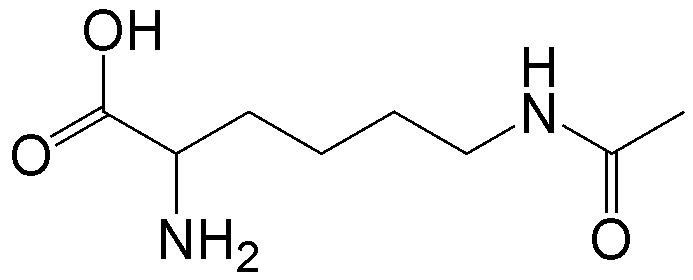
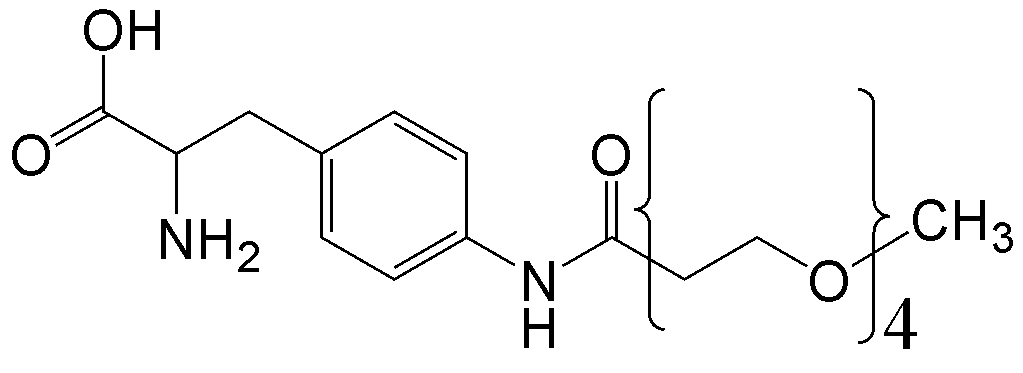
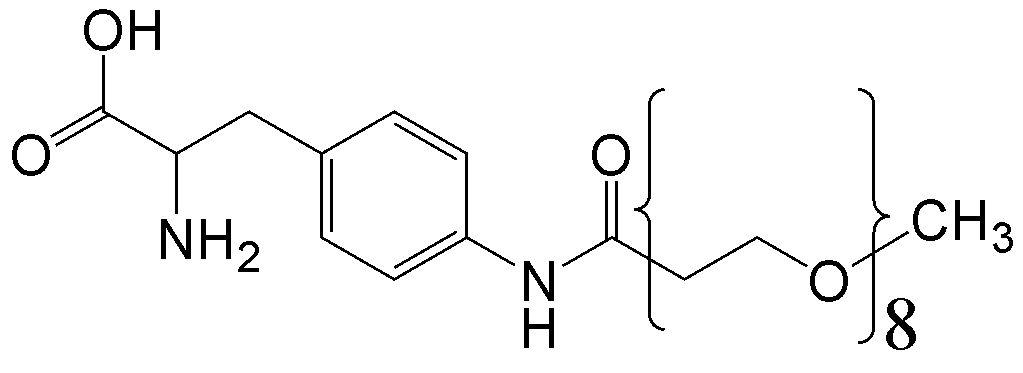
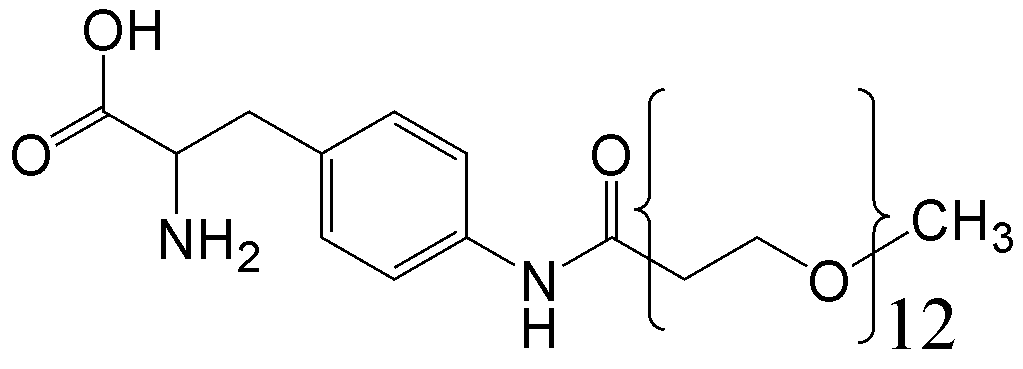
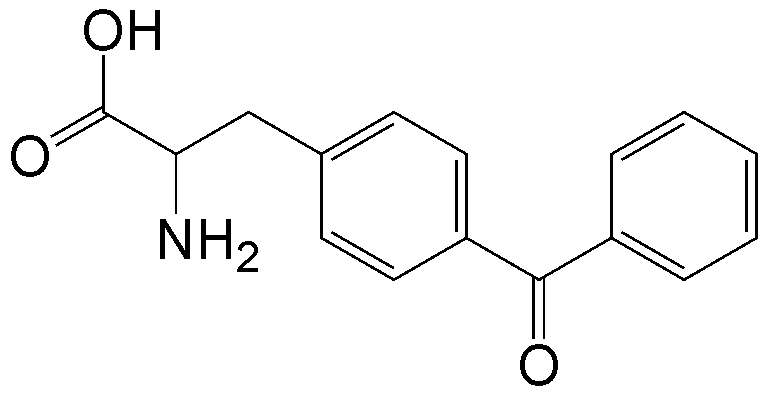
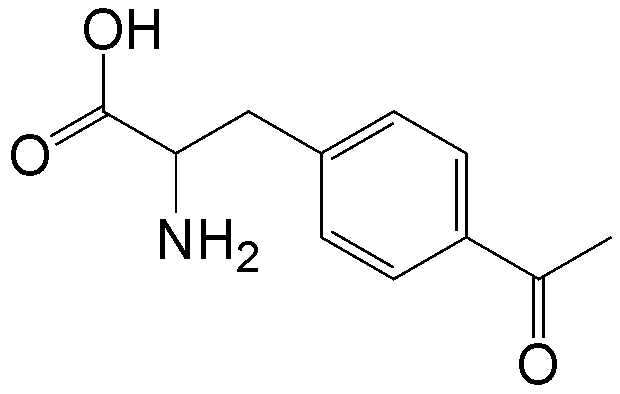
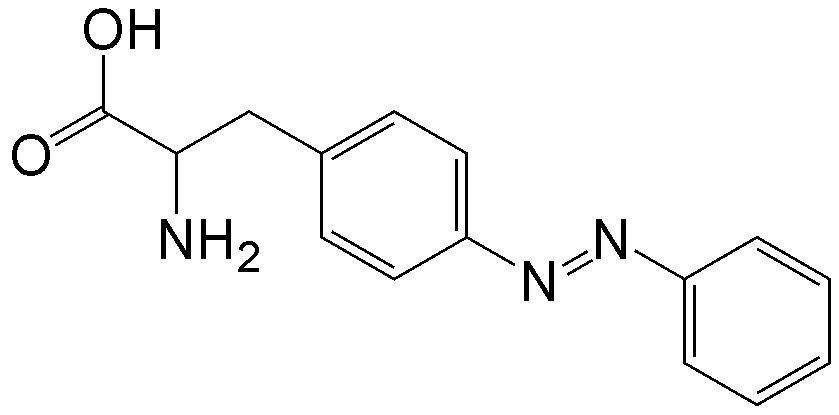
概要
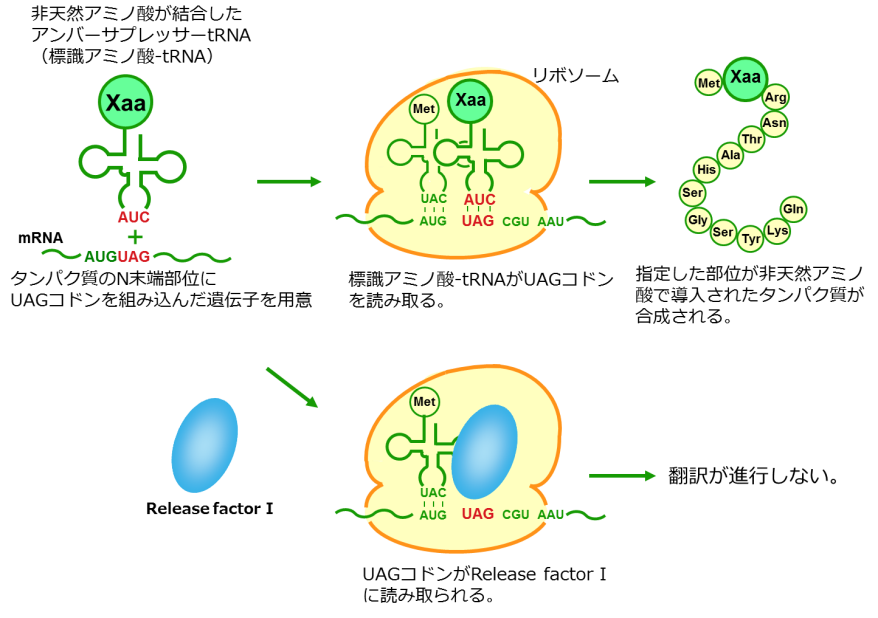
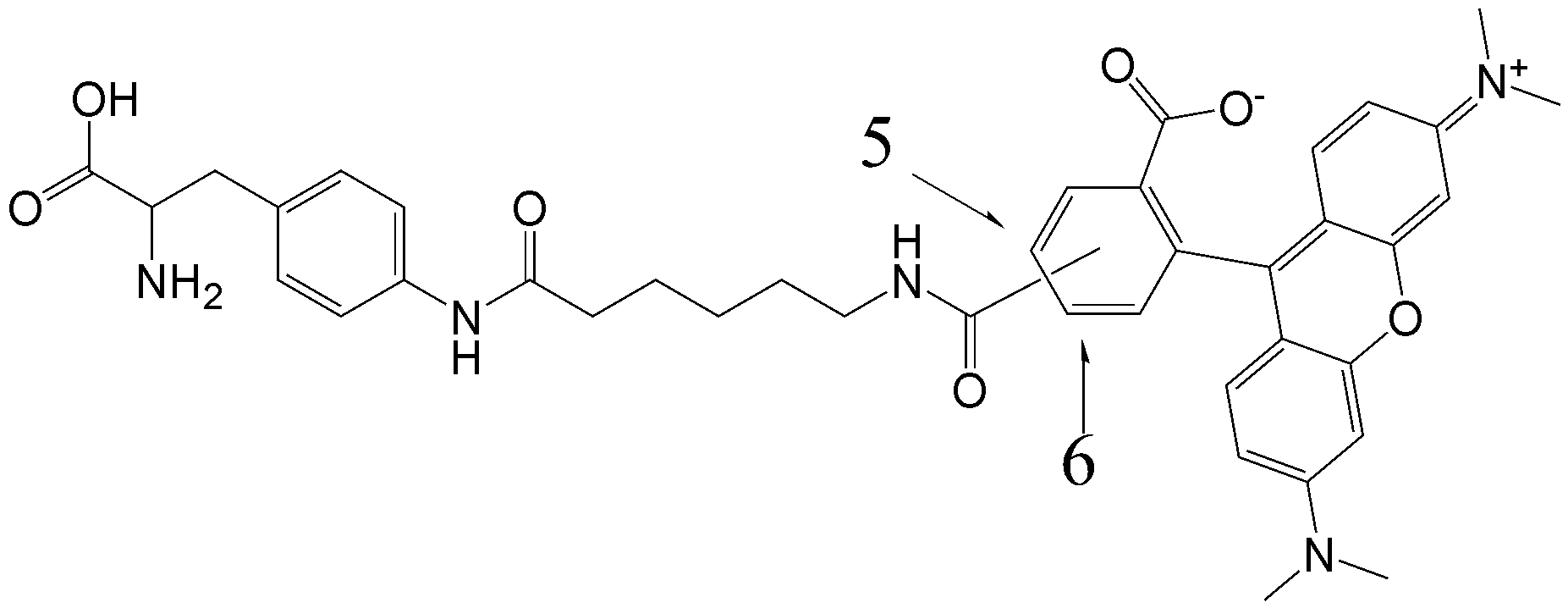
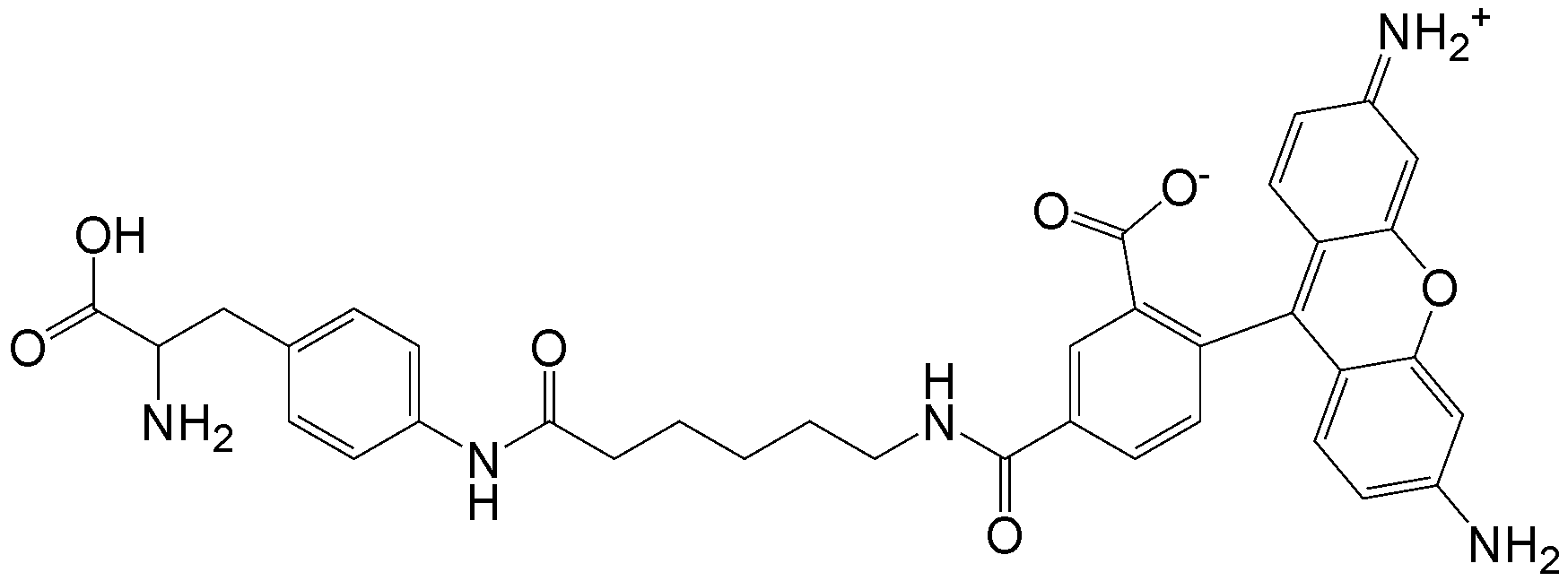
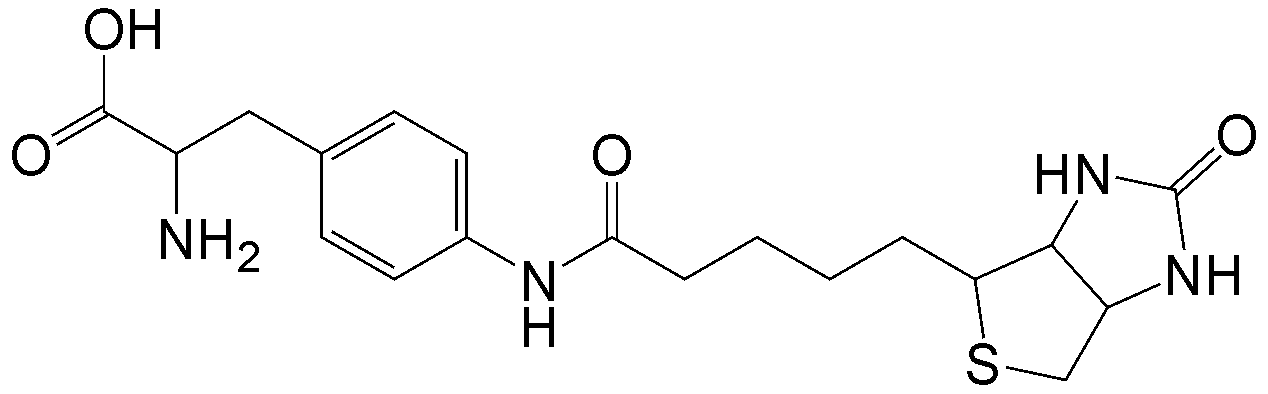
CloverDirect tRNA Reagents for Site-Directed Protein Functionalizationは、無細胞翻訳系を利用してタンパク質の指定した部位に非天然アミノ酸を導入するための試薬です。蛍光基、ビオチン、PEG鎖、架橋剤を側鎖に持つ非天然アミノ酸や、翻訳後修飾アミノ酸などが用意されています。終止コドンの1つであるアンバーコドンまたは4塩基コドンを組み込んだ遺伝子と本製品を無細胞翻訳系に加えるだけで、非天然アミノ酸導入タンパク質を短時間で効率よく合成することができます。
製品リスト
※1 : 導入可能部位
非天然アミノ酸の種類によって、導入可能な部位が異なります。
○ : 導入可能
△ : 部位によっては導入が可能
× : 導入は困難
製品リスト(開始コドン用; CloverDirect initiator)
大腸菌fMet-tRNAのアンチコドンをアンバーアンチコドンに変更し、アミノアシル化した製品です。タンパク質のN末端にクロロアセチル基を保有したアミノ酸を導入することができます。
| コード | 非天然アミノ酸 | コドン | 容量
(µL翻訳反応) |
価格 |
|---|---|---|---|---|
| CLDi-001 | Cl-AcAla [N-Chloroacetyl-DL-alanine] 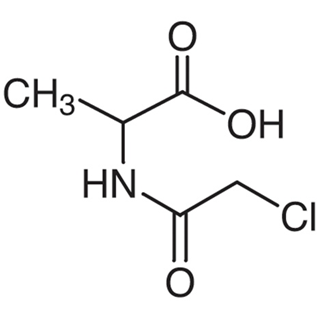 |
amber | 300 | ¥80,000 |
| CLDi-002 | Cl-AcPhe [N-Chloroacetyl-DL-Phenylalanine] 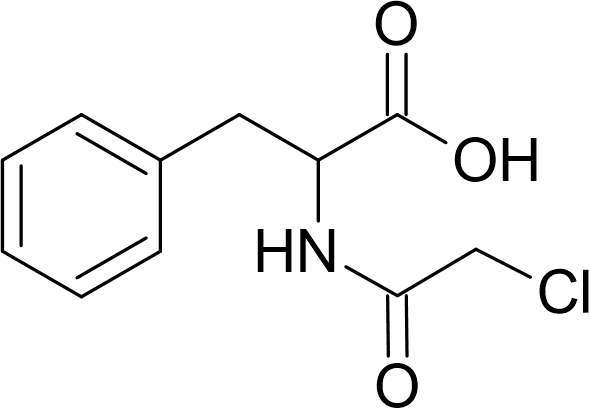 |
amber | 300 | ¥80,000 |
参考文献
1) FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids
Daisuke Kajihara, Ryoji Abe, Issei Iijima, Chie Komiyama, Masahiko Sisido, Takahiro Hohsaka
Nature Methods, 3, 923-929 (2006).
2) Position-specific incorporation of biotinylated non-natural amino acids into a protein in a cell-free translation system
Takayoshi Watanabe, Norihito Muranaka, Issei Iijima, and Takahiro Hohsaka
Biochem. Biophys. Res. Commun., 361, 794-799 (2007).
3) Comprehensive screening of amber suppressor tRNAs suitable for incorporation of non-natural amino acids in a cell-free translation system
Hikaru Taira, Yosuke Matsushita, Kenji Kojima, Kaori Shiraga, Takahiro Hohsaka
Biochem. Biophys. Res. Commun., 374, 304-308 (2008).
4) Efficient Incorporation of Nonnatural Amino Acids with Large Aromatic Groups into Streptavidin in In Vitro Protein Synthesizing Systems
Takahiro Hohsaka, Daisuke Kajihara, Yuki Ashizuka, Hiroshi Murakami, and Masahiko Sisido
J. Am. Chem. Soc., 121, 34-40 (1999).
5) 無細胞翻訳系における非天然アミノ酸の導入技術の開発とその応用
芳坂 貴弘, 生化学, 72(3), 247-253 (2007).
6) 遺伝暗号を拡張した人工タンパク質合成系の開発と応用
芳坂 貴弘, 生物物理, 47(2), 124-128 (2007).
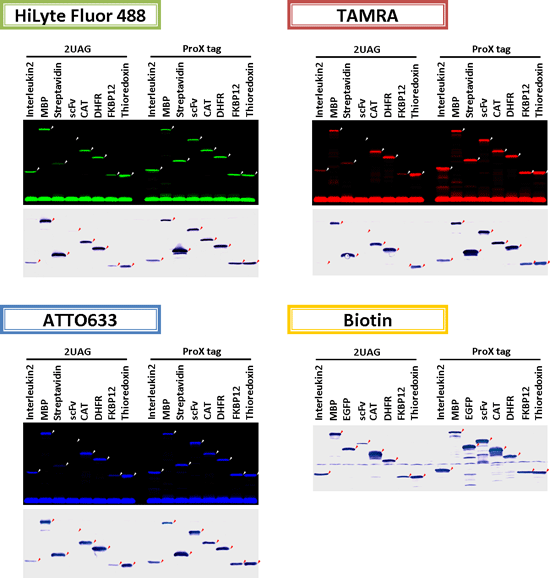
アプリケーションデータ
ピンポイント標識タンパク質の発現例(大腸菌無細胞翻訳系)
製品使用例
1) A protease inhibitor discovery method using fluorescence correlation spectroscopy with position-specific labeled protein substrates.
Nakata H, Ohtsuki T, Sisido M.
Anal Biochem. 2009 Jul 15;390(2):121-5.
2) “Quenchbodies”: quench-based antibody probes that show antigen-dependent fluorescence.
Abe R, Ohashi H, Iijima I, Ihara M, Takagi H, Hohsaka T, Ueda H.
J Am Chem Soc. 2011 Nov 2;133(43):17386-94.
3) Dynamically varying interactions between heregulin and ErbB proteins detected by single-molecule analysis in living cells.
Hiroshima M, Saeki Y, Okada-Hatakeyama M, Sako Y.
Proc Natl Acad Sci U S A. 2012 Aug 28;109(35):13984-9.
4) Development of a novel PPARγ ligand screening system using pinpoint fluorescence-probed protein.
Nagai H, Ebisu S, Abe R, Goto T, Takahashi N, Hosaka T, Kawada T.
Biosci Biotechnol Biochem. 2011;75(2):337-41.
5) Ultra Q-bodies: quench-based antibody probes that utilize dye-dye interactions with enhanced antigen-dependent fluorescence.
Abe R, Jeong HJ, Arakawa D, Dong J, Ohashi H, Kaigome R, Saiki F, Yamane K, Takagi H, Ueda H.
Sci Rep. 2014 Apr 11;4:4640.
6) Strategy for making a superior Quenchbody to proteins: effect of the fluorophore position.
Jeong HJ, Ueda H.
Sensors (Basel). 2014 Jul 23;14(7):13285-97.
7) In vitro selection of a photoresponsive peptide aptamer to glutathione-immobilized microbeads.
Tada S, Zang Q, Wang W, Kawamoto M, Liu M, Iwashita M, Uzawa T, Kiga D, Yamamura M, Ito Y.
J Biosci Bioeng. 2015 Feb;119(2):137-9.
8) A fluorogenic peptide probe developed by in vitro selection using tRNA carrying a fluorogenic amino acid.
Wang W, Uzawa T, Tochio N, Hamatsu J, Hirano Y, Tada S, Saneyoshi H, Kigawa T, Hayashi N, Ito Y, Taiji M, Aigaki T, Ito Y.
Chem Commun (Camb). 2014 Mar 18;50(22):2962-4.
9) Genetic PEGylation.
Tada S, Andou T, Suzuki T, Dohmae N, Kobatake E, Ito Y.
PLoS One. 2012;7(11):e49235.
10) In vitro selection of a photo-responsive peptide aptamer using ribosome display.
Liu M, Tada S, Ito M, Abe H, Ito Y.
Chem Commun (Camb). 2012 Dec 18;48(97):11871-3.
11) A cell-free translocation system using extracts of cultured insect cells to yield functional membrane proteins.
Ezure T, Nanatani K, Sato Y, Suzuki S, Aizawa K, Souma S, Ito M, Hohsaka T, von Heijine G, Utsumi T, Abe K, Ando E, Uozumi N.
PLoS One. 2014 Dec 8;9(12):e112874.
12) Farnesyl pyrophosphate regulates adipocyte functions as an endogenous PPARγ agonist.
Goto T, Nagai H, Egawa K, Kim YI, Kato S, Taimatsu A, Sakamoto T, Ebisu S, Hohsaka T, Miyagawa H, Murakami S, Takahashi N, Kawada T.
Biochem J. 2011 Aug 15;438(1):111-9.
13) Detection method for quantifying global DNA methylation by fluorescence correlation spectroscopy
Tomohiro Umezu, Kazuma Ohyashiki, Junko H Ohyashiki
Anal Biochem. 2011 Aug 15;415(2):145-50.
14)Autonomous synthesis and assembly of a ribosomal subunit on a chip
Michael Levy, Reuven Falkovich, Shirley S Daube, Roy H Bar-Ziv
Sci Adv. 2020 Apr 15;6(16):eaaz6020.
15)Reconstitution of Bam Complex-Mediated Assembly of a Trimeric Porin into Proteoliposomes
Sunyia Hussain, Janine H Peterson, Harris D Bernstein
mBio. 2021 Aug 31;12(4):e0169621.
16)Genetic Code Expansion and a Photo-Cross-Linking Reaction Facilitate Ribosome Display Selections for Identifying a Wide Range of Affinity Peptides
Takuto Furuhashi, Kensaku Sakamoto, Akira Wada
Int J Mol Sci. 2023 Oct 27;24(21):15661.
技術資料
1.アプリケーションデータCLD-001(PDF、2018/9/10追加);非天然アミノ酸導入例1
2.アプリケーションデータCLD-002(PDF、2025/1/22追加);非天然アミノ酸導入例2
3.アプリケーションデータCLD-003(PDF、2025/10/29追加);非天然アミノ酸導入例3
4.アプリケーションデータCLD-004(PDF、2025/10/31追加);非天然アミノ酸導入例4, 開始コドンへの導入